Release January 5th, 2024
New ColorMIP Matching Widget
A new ColorMIP matching widget enables matching of Genetic Lines-to-EM neurons, EM-to-EM neurons, and EM Neurons-to-Genetic Lines with a few seconds response time of a few seconds across 100 collections and more than 800k ColorMIP images in total. Horizontal mirror of the search images allows to find contralateral neurons. The matching widget not only produces matches for candidate genetic lines, but it works also nicely to find morphologically similar neurons in other datasets (especially useful for proofreading neurons by finding completed cells in other datasets for the same celltype).
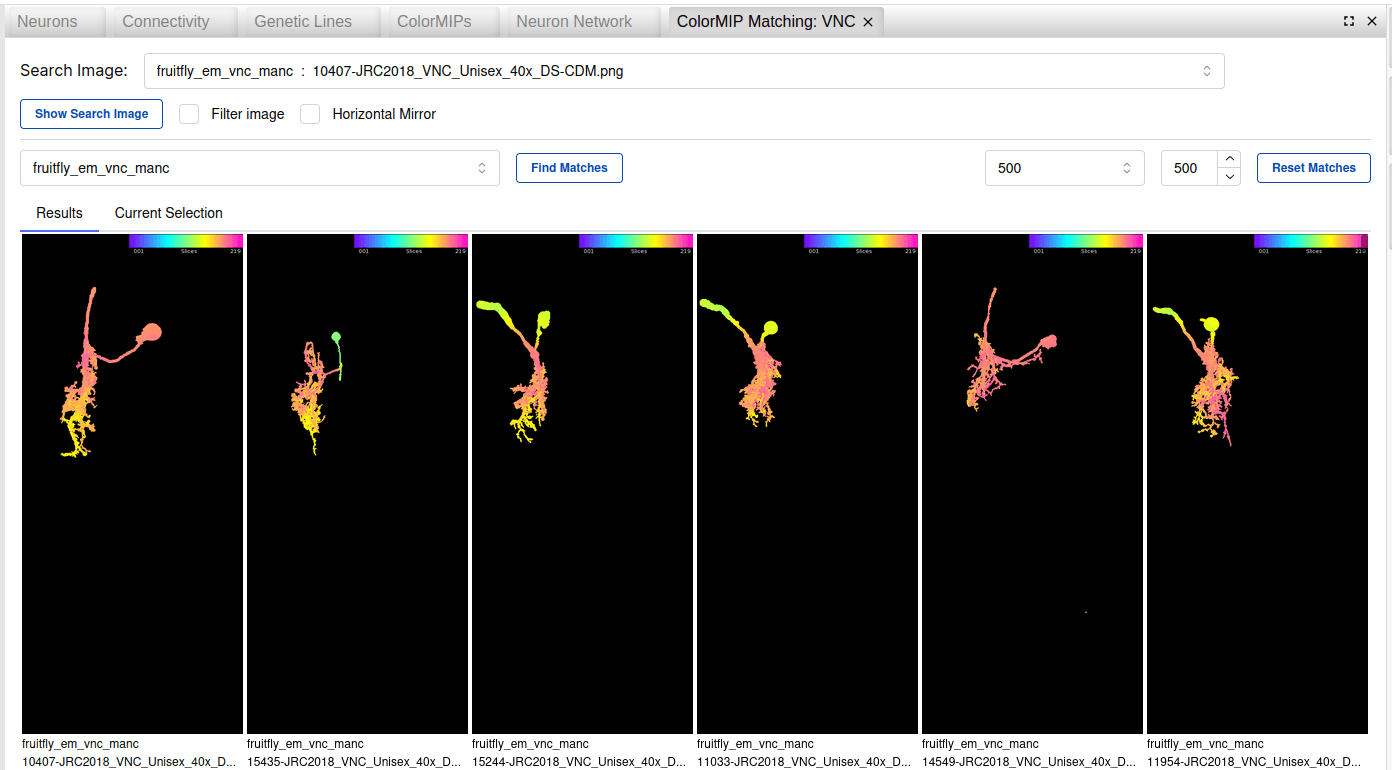
In order to map EM neurons from published or community (CAVE-backed) datasets, you can use the ColorMIPs widget to select the dataset and provide a set of segmentIDs to retrieve precomputed (for published) and on-the-fly generated (for community) ColorMIPs. You can then select the ColorMIPs you want to match, which opens a new widget for the mapping procedure where you can specify the target collection (single collections, or aggregate collections for all EM or LM images for either brain or VNC).
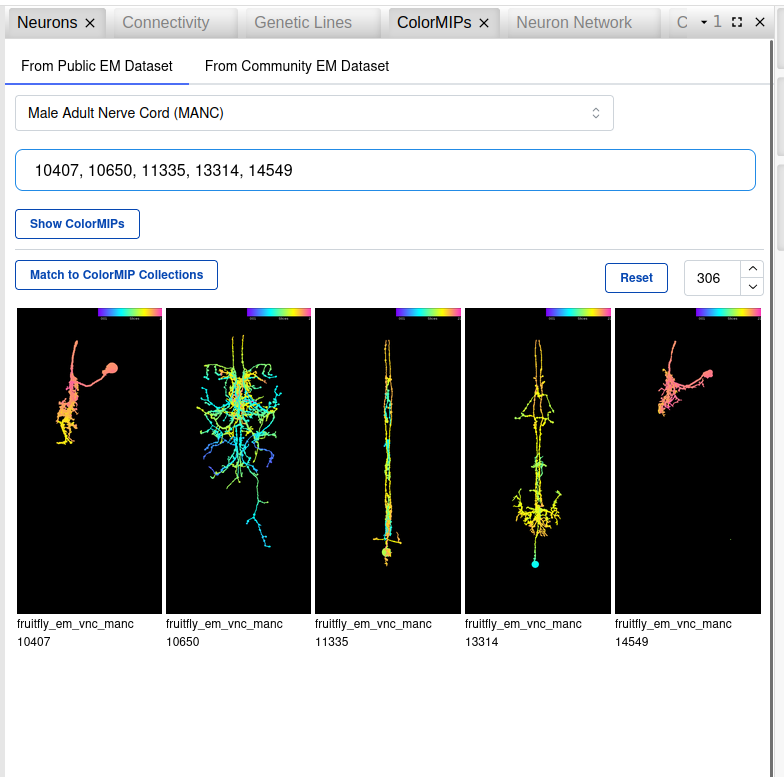
In order to map genetic lines, you can use the new Genetic Lines widget which allows you to search for a line name and retrieve all available ColorMIPs from the genetic lines collection. You can then select the the images you want to map. You can then select the images of interest for matching.
Please make sure to cite the appropriate publications when you use ColorMIP from either EM neurons or genetic lines in your work. The genetic lines ColorMIPs are obtained from Janelia Research Campus FlyLight project. Please look at the following list for appropriate references:
Links to references for neurons in published EM datasets can be found in the About widget (e.g. for FlyWire, FANC, MANC).
- NeuronBridge
- Janelia FlyLight Imagery 1
- Janelia FlyLight Imagery 2,
- Janelia Flylight Expression Patterns of GAL4 and LexA Driver Lines
- Fly Light Split-GAL4 Driver Collection
- Also check the latest news on the Janelia FlyLight website for recent publications and links to fruitfly stock facilities.
Links to references for neurons in published EM datasets can be found in the About widget (e.g. for FlyWire, FANC, MANC).
New Human cerebral cortex (H01) dataset
The petascale human cortex dataset has been added. The 46k neurons and their metadata can be browsed from the Neurons widget, and connectivity information is available from the Connectivity widget.
Release September 4th, 2023
New Neuron Widget
Public datasets contain now a new Neuron widget (replacing the Segment widget) with an improved table user interface with global search, per-column filtering, reordering of columns etc. Individual entries can be copied to the clipboard on click (useful e.g. for segment ID column).
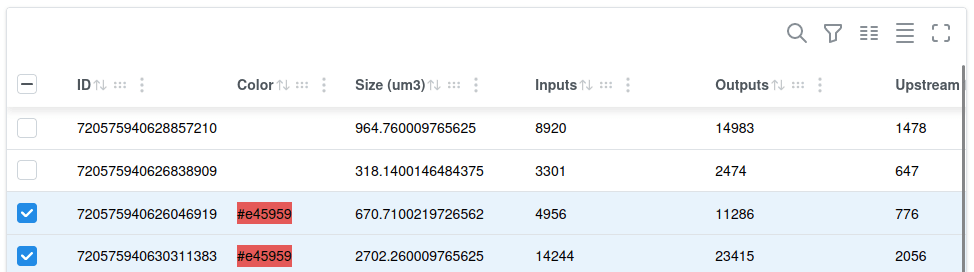
ColorMIP Atlas for FANC
More than 18k colorMIP images of FANC neurons are now downloadable (only with community access) from the About widget.

Release August 15th, 2023
Cell Type Atlas
First version of a cell type atlas with underlying data model. Individual proofread neurons in specific datasets have to be mapped to commonly agreed cell type identifiers in a specific species according to a particular author or publication. This cell type atlas uses this data model to represent all this information to map reconstructed public neurons from EM datasets to cell types in different species, starting with fruit flies. The interfaces provides an entry point to filter cell types in a species, and potentially also brain region, and lists all the unique identifiers for cells in any of the datasets available on the platform. These identifiers can be used as entry points to retrieve the neurons, and further study their morphology, connectivity or other annotations available.
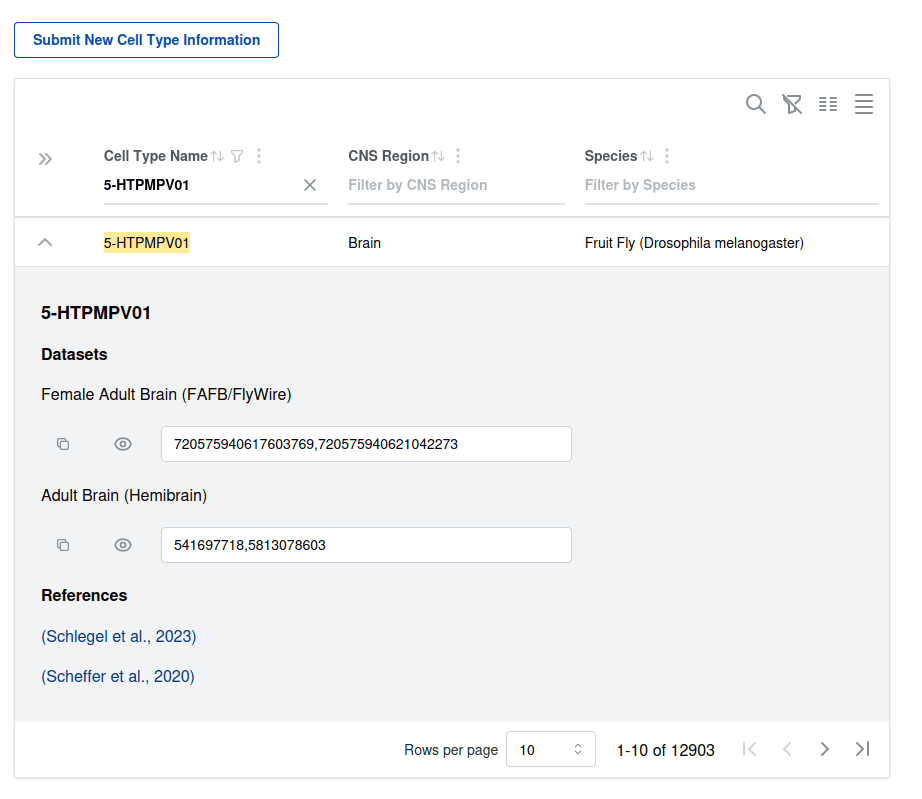
It is important to note that each definition of a cell type identifier, or each statement that a particular neuron in a specific dataset is an instance of a cell type, has to be supported by an author or publication. Thus it is possible to have mutually exclusive or overlapping cell typing nomenclatures or schemes for any given datasets. We do not attempt to curate and merge such different nomenclature. We do assume that the same cell type identifiers (exact spelling) in different nomenclatures corresponds to the same criteria used to define the cell type.
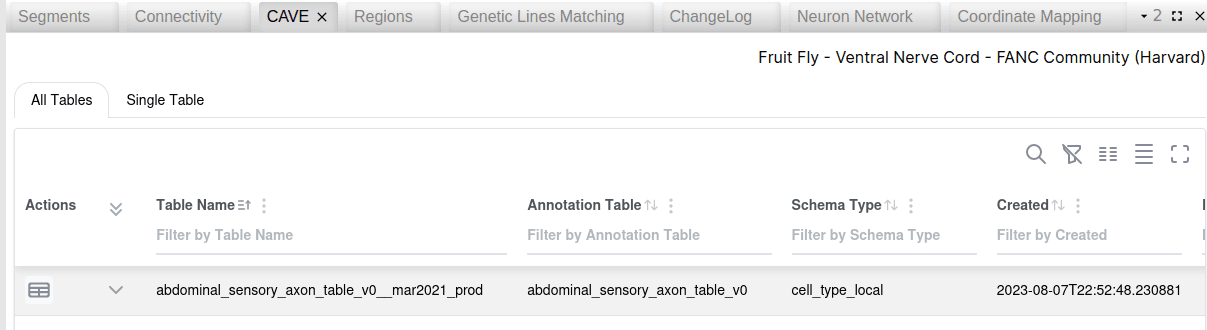
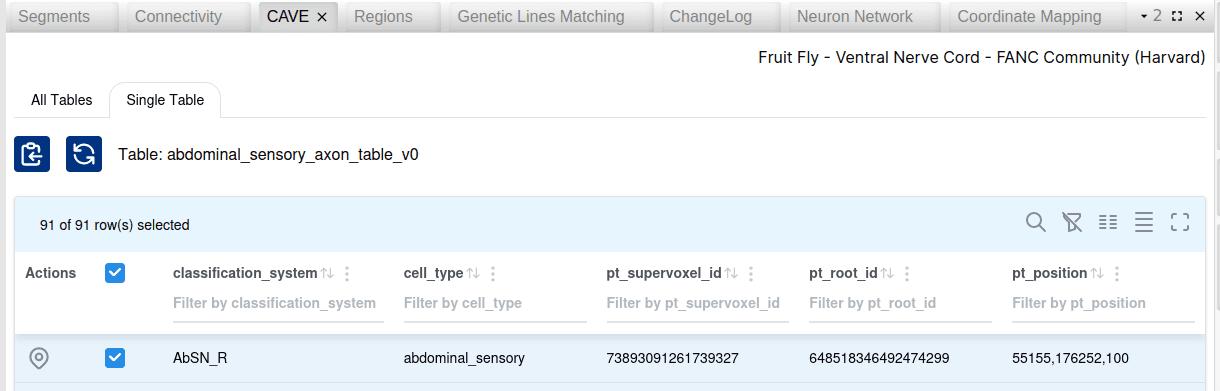
CAVE Table Widget
A new CAVE table widget allows for datasets that are backed by the CAVE infrastructure to retrieve a list of all tables with their description. Individual tables can be opened and their records are retrieved in a table. For annotations with an associated spatial location, a button allows to navigate to the 3d location in Neuroglancer. Records can be filtered and selected, and their corresponding segment IDs copied for futher usage in other widgets.
Publication Feed
The new Feed section of the website collects recent publications in the field of connectomics, covering all stages of the data generation pipeline as well as applications to address biological circuit questions, and review and methods publications. The entries do not need to contain a link to a peer-reviewed publication, but can also link to tweets, or any other web resources. The hashtags help with finding useful entries and may later be used for filtering. One goal is to highlight how neurons and circuits mapped based on electron microscopy have been useful to uncover principles of neural computation.
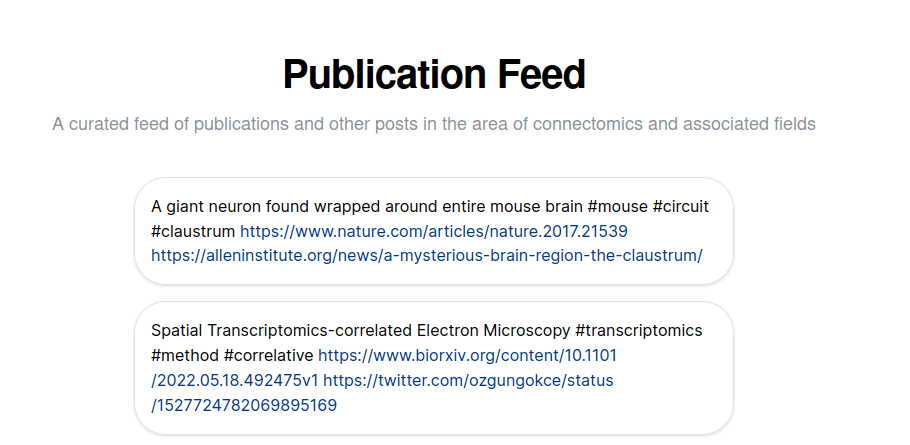
The feed is generated from a user of the decentralized social network Nostr. Specifically, from user account. If you like to contribute to this feed, please get in touch. You can simply setup an anonymous user account on Nostr.rocks and share the public key with us. If the feed is appropriate and relevant, we are happy to include it here.
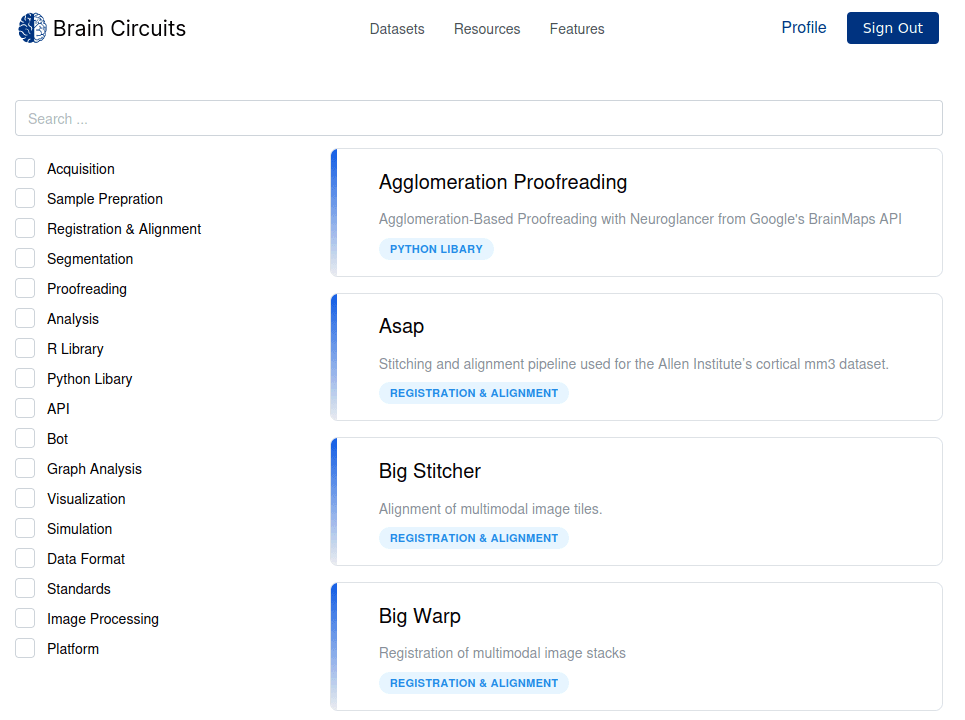
Tools and Resources
A new interface for the Tools and Resources section allows to do full-text search across record titles, descriptions and tags. Each record can also be filtered by its corresponding tags. This allows to quickly find and explore resources that may be relevant for a particular stage in the connectomics pipeline.
Previous Releases
Datasets
Integrated public datasets:
- Fruitfly VNC - MANC Dataset: 23k proofread neurons and connectivity
- Fruitfly Brain - FAFB/FlyWire Dataset: 128k proofread neurons and connectivity
- Fruitfly Brain - Hemibrain Dataset: 25 proofread neurons and connectivity